Welcome to the Interactive Pathways Explorer v2
interactive Pathways Explorer (iPath) is a web-based tool for the visualization, analysis and customization of the various pathways maps. Current version provides three different global overview maps:
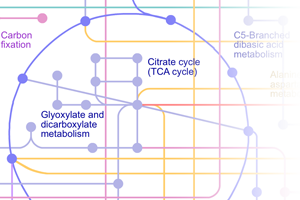
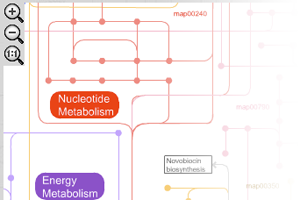
- Metabolic pathways: constructed using 146 KEGG pathways, and gives an overview of the complete metabolism in biological systems
- Regulatory pathways: 22 KEGG regulatory pathways
- Biosynthesis of secondary metabolites: contains 58 KEGG pathways involved in biosynthesis of secondary metabolites
In addition to the KEGG based overview maps, iPath is used for visualization of various species specific, manually created pathway maps. At the moment, only Mycoplasma pneumoniae and a beta version of Mycobacterium tuberculosis maps are publicly available.
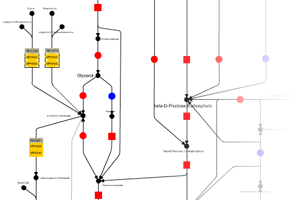
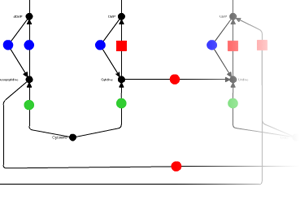
iPath provides extensive map customization and data mapping capablities. Colors, width and opacity of any map element can be customized using various types of data (for example KEGG KOs, COGs or enzyme EC numbers). Our help pages provide the full list of supported data types and required formats. All maps in iPath can be easily converted to various bitmap and vector graphical formats for easy inclusion is your documents or further processing.
If you find iPath useful in your work, you can cite
- Letunic et al. (2008) Trends Biochem Sci. 33(3):101-3 iPath: interactive exploration of biochemical pathways and networks. (PDF)
- Yamada et al. (2011) Nucleic Acids Res. 39(suppl 2): W412-W415 iPath2.0: interactive pathway explorer. (PDF)