Help and FAQs
We tested the following:
- Linux/Unix: Google Chrome, Firefox and Opera work fine with Flash 10 plugin.
- Windows: Microsoft Internet Explorer 7 or later,Google Chrome, Firefox and Opera work fine.
- Mac: Google Chrome, Firefox, Opera and Safari all work without problems.
Certain features of iPath (restoring selections and access to previously exported files) require access to Flash shared storage ('Flash cookies').
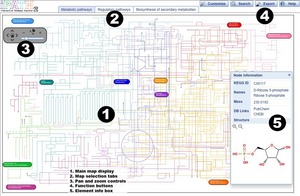
Overview
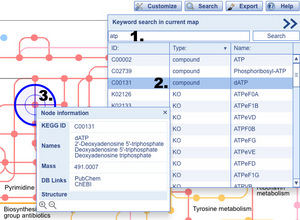
Search
Basics
|
iPath2 provides powerful data mapping tools and extensive customization options. Clicking the 'Customization' button will open a panel (shown on the right) which provides access to all available customization options. Hover your mouse cursor over individual options to show a short explanation in a popup balloon. Various options are described in detail below:
|
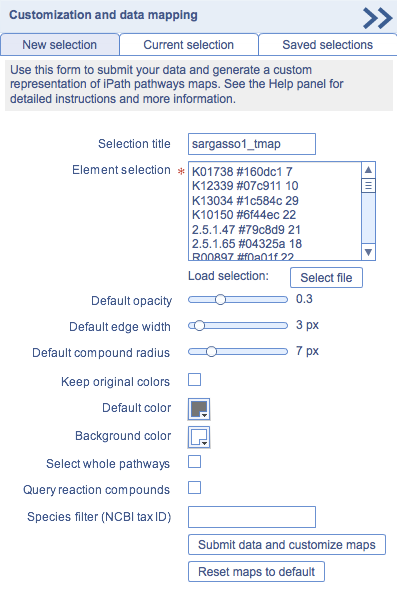 Customization panel |
Supported data types
The following data types are supported by iPath, and can be used to customize the maps. Make sure you use the required prefix for each data type, as shown in the examples below.
| Data type | Prefix | Example ID |
|---|---|---|
| KEGG Pathways | - | 00650 |
| KEGG Compounds | - | C00003 |
| KEGG KOs | - | K01000 |
| STRING proteins | - | 224324.AQ_626 |
| KEGG proteins | - | aae:aq_626 |
| COGs/eggNOGG OGs | - | COG0007 |
| Enzyme EC numbers | E or EC | E2.4.1.82 |
| Uniprot IDs/ACCs | UNIPROT: | UNIPROT:Q93015 |
| IPI IDs | - | IPI00745889 |
| NCBI GI IDs | ncbi-gi: | ncbi-gi:326314893 |
Current selection's map matching statistics
Saving and restoring element selections
If a Selection title is specified when submitting data, all selection parameters will be stored on the server, and can be simply accessed through the "Saved selections" tab. Use the icons next to each saved selection name to either restore the selection, or delete it.
Example dataset
An example dataset file is available for download, which maps the enzymatic activity in the human gut microbiome (metagenomic data derived from Japanese fecal samples). First, second and third columns represent 1:ortholog ID in KEGG, 2:abundance of the OG (represented using element width, 'W') and 3:color in the map respectively. Simply paste the contents of the file into iPath's element selection box, and set map background color to black for best results.
Customized maps in iPath can be saved in various output formats. Clicking the 'Export' button will open the export panel, shown on the right. Maps can be exported into any of the following formats:
Note: Due to technical limitations, transparency cannot be preserved in EPS and PDF formats. If you provide an optional export title, your exported map will be stored on the server for 30 days, and accessible through the 'Saved exports' tab. |
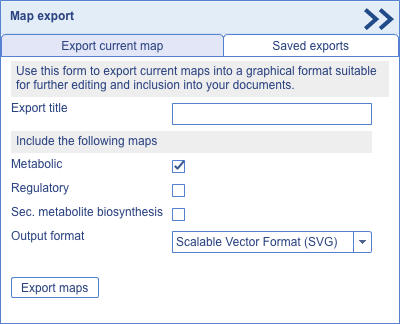 Map export panel |