Welcome to the Interactive Pathways Explorer
This version of iPath is now outdated. Please visit http://pathways.embl.de to access the latest version.
If you find iPath useful in your work, you can cite Letunic et al. (2008) Trends Biochem Sci. 33(3):101-3 iPath: interactive exploration of biochemical pathways and networks. (PDF)
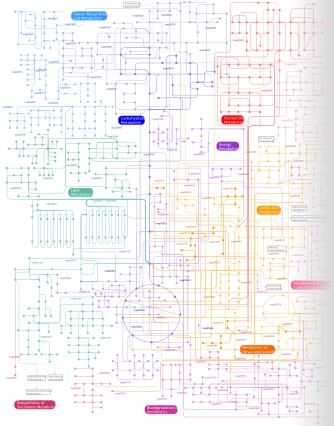 Click on the image to display the interactive version |
Global map of metabolic pathways
The interactive viewer provides an easy way of navigating the global pathways map.
How to navigate the map:
Clicking on any node or edge in the map will provide detailed information, including links to the relevant KEGG annotation pages. In addition to the interactive version, map can be downloaded in the following graphical formats:
|
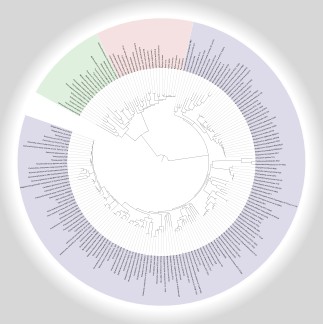 Click on the image to display the interactive version. |
In addition to the default global map which includes data from all species in KEGG, iPath provides complete metabolic pathways maps for individual species and various taxnomic classes. Clicking on the tree image on the left will display an interactive phylogenetic tree, generated using iTOL. You can navigate the tree using the same shortcuts which are used in the map view (mousewheel, arrows, Page Up, Page Down and Home). In addition, you can search the tree for any species or taxnomic class. The tree includes 183 species (21 Eukayota, 144 Bacteria and 18 Archaea). Click on any species or clade in the tree to display the links to the relevant pathways map. There are two versions of each map available:
This tool allows easy comparison of metabolic pathway maps for multiple species or classes. |